About NWB

Progress in modern science is enabled by data sharing. There are major obstacles, however, that limit the open exchange of data particularly in neuroscience, a field where many small laboratories are pursuing diverse questions about the brain using a greatvariety of tools and techniques. In mid 2014, The Kavli Foundation initiated Neurodata Without Borders, a consortium of researchers and foundations with a shared interest in breaking down the obstacles to data sharing.
Overview
The aim of Neurodata Without Borders is to standardize neuroscience data on an international scale. Our goal is to break down the geographic, institutional, technological and policy barriers that impede the flow of neuroscience data within the greater scientific community. Our intent is to accelerate the pace of discovery and success of brain research worldwide.
The need to standardize neuroscience data has never been more urgent. Large-scale brain research projects such as the U.S. BRAIN Initiative and resources such as the Allen Cell Types Database are producing massive quantities of data. But the full benefits of these data will not be realized if they cannot easily be shared, pooled and analyzed.

NWB
Unlike in other fields (i.e. genetics and cell biology), neuroscience does not have a standardized way to collect and share the wealth of existing data among researchers. The lack of a common format has made comparison across laboratories difficult and replication of specific experiments almost impossible, significantly slowing overall progress in the field.
Neurodata Without Borders (NWB) is a data standard for neurophysiology, providing neuroscientists with a common standard to share, archive, use, and build common analysis tools for neurophysiology data. NWB is designed to store a variety of neurophysiology data, including data from intracellular and extracellular electrophysiology experiments, data from optical physiology experiments, and tracking and stimulus data. The project includes not only the NWB format, but also a broad range of software for data standardization and application programming interfaces (APIs) for reading and writing the data as well as high-value datasets that have been translated into the NWB data standard.
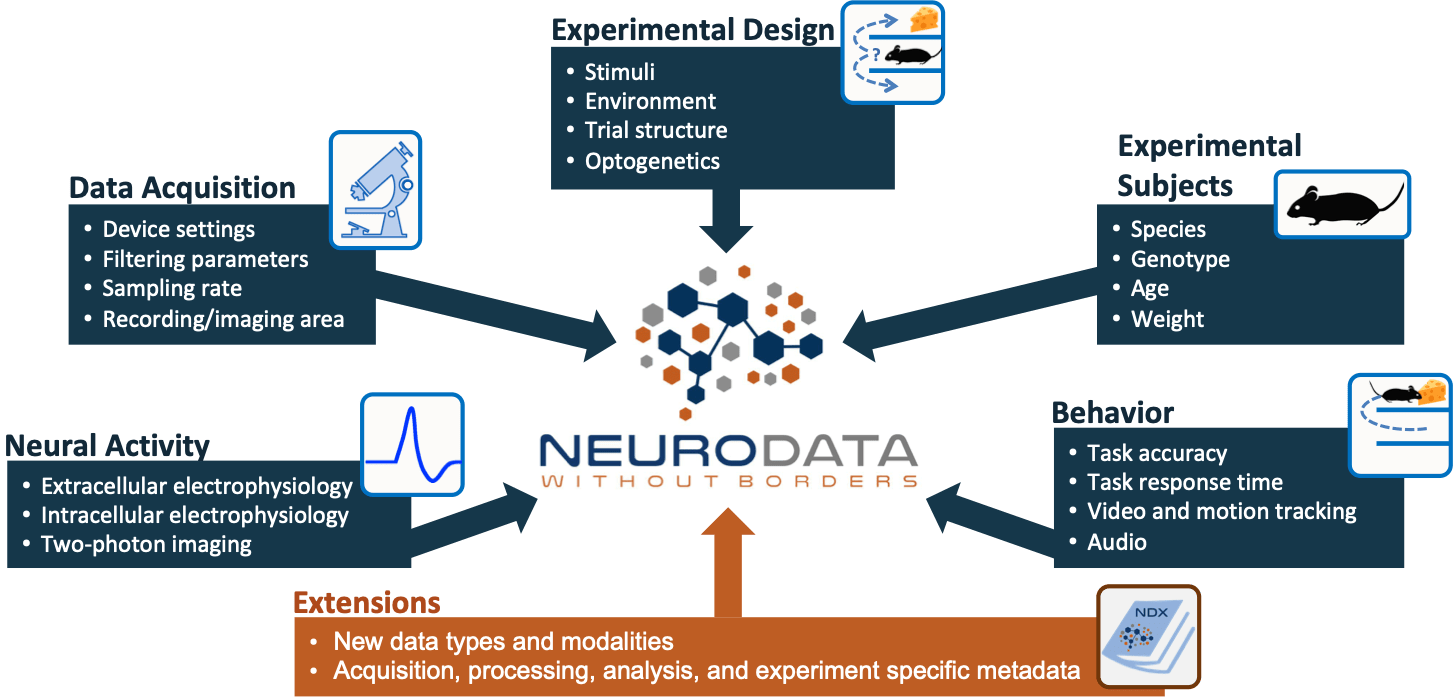
Neurodata Without Borders is intended to serve the broad neuroscience community and encourage the sharing of data by scientists worldwide. NWB 2.0 was released in February 2019. Please give it a try. Join our mailing list for updates or ask questions on our Slack channel.
To learn more about the approach taken to develop the NWB Format, please read our open access eLife article.
Technical Design Goals of the NWB Format
 Cross-platform
Cross-platform Support for tool makers
Support for tool makers Quickly develop a basic understanding of an experiment and its
Quickly develop a basic understanding of an experiment and its Review an experiment’s details without programming knowledge
Review an experiment’s details without programming knowledge Accommodate an experiment’s raw and processed data
Accommodate an experiment’s raw and processed data Encapsulate all of an experiment’s data, or link to external data sources when necessary
Encapsulate all of an experiment’s data, or link to external data sources when necessary Data published in the format should be accessible for decades
Data published in the format should be accessible for decades Accommodate future experimental paradigms without sacrificing backwards compatibility
Accommodate future experimental paradigms without sacrificing backwards compatibility Support custom extensions when the standard is lacking
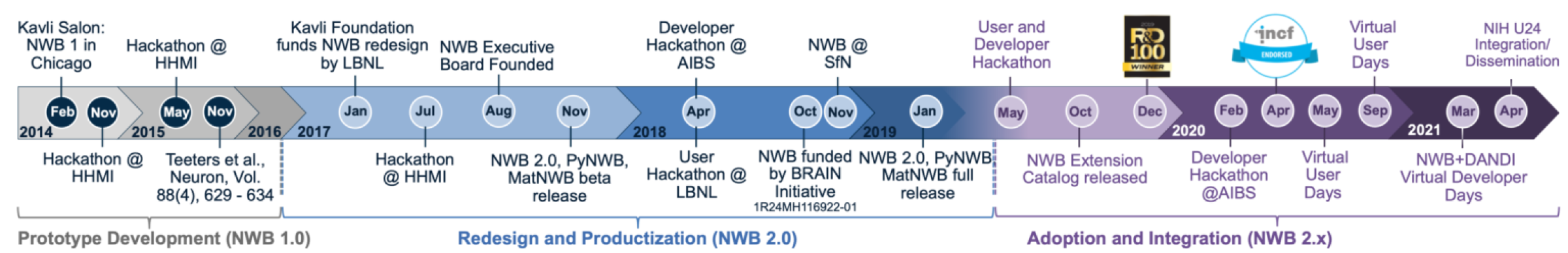
Support custom extensions when the standard is lackingHistory of Neurodata Without Borders (NWB)
Origins and NWB 1.0 (2014-2016)
Neurodata Without Borders: Neurophysiology was launched in mid-2014 as a collaborative initiative to develop a unified data format for cellular-based neurophysiology data. The Kavli Foundation played an important role in initiating this movement, bringing together key stakeholders to address the growing challenge of data interchange between laboratories.
The initial development of NWB 1.0 was funded by the Kavli Foundation along with other industry partners and private foundations, with scientific leadership from key neuroscience laboratories: the Allen Institute for Brain Science (AIBS), the Svoboda Lab (Janelia Research Campus), the Meister Lab (Caltech), the Buzsáki Lab (NYU), and Fritz Sommer/Jeff Teeters (UC Berkeley, maintainers of CRCNS.org). This consortium approach ensured the format addressed real-world use cases from diverse experimental paradigms. The first hackathon at Janelia initiated this collaborative effort.
After an intensive one-year development cycle that included extensive testing with representative datasets, NWB 1.0 was released in 2015 as the first standardized format specifically designed for neurophysiology data. This work was showcased at the second hackathon at Janelia.
Evolution to NWB 2.0 (2017-2019)
Building on lessons learned from NWB 1.0 adoption, the project entered a second phase with expanded leadership including Dr. Kris Bouchard, Dr. Oliver Rübel (Lawrence Berkeley National Laboratory, LBNL), Dr. Loren Frank (UCSF), and Dr. Edward Chang (UCSF). This phase began with the third hackathon at Janelia and marked a significant shift in the project’s approach, focusing on:
- Creating a robust software architecture to support the data standard
- Developing a comprehensive standards ecosystem rather than just a file format
- Establishing an open community development model
- Refining the core data standard based on community feedback
The technical redevelopment of NWB 2.0 was led by scientific software engineers at LBNL, principally Dr. Oliver Rübel and Andrew Tritt, working in close collaboration with neuroscience laboratories and the broader community. This collaboration ensured the format remained scientifically relevant while gaining stronger software engineering foundations. A series of hackathons (Seattle 2018, Berkeley 2018) helped drive this development.
NWB 2.0 was officially released in February 2019, featuring a completely redesigned architecture based on modern software development practices, enhanced extensibility through schema language, improved storage efficiency, and a comprehensive Python API (PyNWB) and MATLAB API (MatNWB) for programmatic interaction with NWB files. The Janelia 2019 hackathon celebrated this milestone release.
Community Growth and Governance (2019-Present)
Following the release of NWB 2.0, the project established a formal governance structure to ensure sustainability and community-driven development. This structure consists of:
- An Executive Board (EB) that guides the overall vision, strategic roadmap, fundraising, and outreach
- A Technical Advisory Board (TAB) that makes technical decisions and maintains the software ecosystem
- Working groups focused on specific aspects of the standard and its implementation
The NWB ecosystem has continued to expand with additional tools including:
- Neurosift, NWB Widgets and NWB Explorer for interactive visualizations of NWB data
- Integration with analysis frameworks like CaImAn, suite2p, and SpikeInterface
- Data validation tools such as NWB Inspector to ensure format compliance
- Extensions for specialized modalities and experimental paradigms
Adoption and Impact
NWB has gained steady adoption across the neuroscience community. Notable users include the Allen Institute for Brain Science (which has released extensive datasets in NWB format), as well as the Bouchard lab (LBNL/UCB), Svoboda lab, Meister lab, Frank lab (UCSF), Chang lab (UCSF), and many others.
The format now serves as the foundation for major data sharing initiatives, including the BRAIN Initiative archives and the DANDI (Distributed Archives for Neurophysiology Data Integration) project, which leverages NWB as its primary data format. This adoption has facilitated unprecedented data sharing across laboratories, accelerating discovery and collaboration in neurophysiology research. For more information, see our publications and grants and projects.
As the community continues to grow, NWB remains under active development, with regular releases addressing new use cases, experimental modalities, and integration with emerging technologies in neuroscience research. Join us at our upcoming events to learn more and contribute to the project.